Bryson Deanhardt
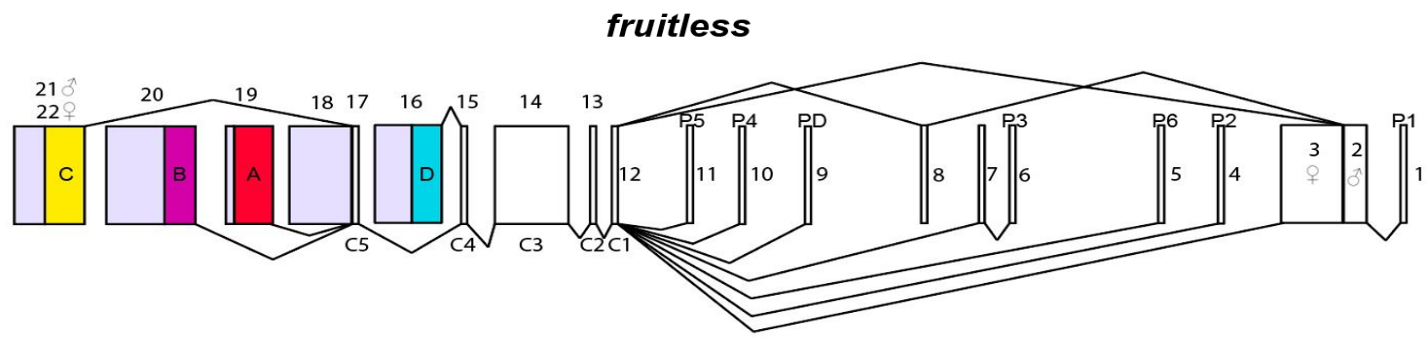 In the animal kingdom, the ability to distinguish environmental and social cues is inherent to the survival and reproduction of a species. In particular, the ability of a species to distinguish and disseminate anatomical and pheromone-based differences that define one species from another. My interest lies at the intersection of two realms, epigenetics and genomics of behavioral circuits as well as evolution and development. In the Jones lab, I explore these two subsections of biology by expanding upon my work at Duke University in the lab of Pelin Volkan on Drosophila melanogaster by exploring the interrelationship of hormone signaling complexes, Methoprene-tolerant (MET) and Germ Cell enhanced (GCE), as well as epigenetic regulation of gene expression and gene splicing around the gene fruitless, a key sex determination and master courtship regulator in insect species. Current research from our lab and others has determined that social experience is regulated by changes in the expression of fruitless through the activation of olfactory sensory neurons in combination with internal hormone states [1,2].
In the animal kingdom, the ability to distinguish environmental and social cues is inherent to the survival and reproduction of a species. In particular, the ability of a species to distinguish and disseminate anatomical and pheromone-based differences that define one species from another. My interest lies at the intersection of two realms, epigenetics and genomics of behavioral circuits as well as evolution and development. In the Jones lab, I explore these two subsections of biology by expanding upon my work at Duke University in the lab of Pelin Volkan on Drosophila melanogaster by exploring the interrelationship of hormone signaling complexes, Methoprene-tolerant (MET) and Germ Cell enhanced (GCE), as well as epigenetic regulation of gene expression and gene splicing around the gene fruitless, a key sex determination and master courtship regulator in insect species. Current research from our lab and others has determined that social experience is regulated by changes in the expression of fruitless through the activation of olfactory sensory neurons in combination with internal hormone states [1,2].
 The genetic network which underlies the changes in behavior also plays a role in the development and specification of olfactory sensory neuron (ORN) receptor expression, ORN number, size, and shape. Previous work on both forelimb and antennae development has shown the necessity of fruitless expression as well as additional factors like Dachshund, Distal-less, and Notch signaling pathways in the regulation of ORN position, antennal development, and segmentation [3,4,5]. Following upon these works current work in the lab aims to expand upon the knowledge of these systems by looking at the association of antennal development across various Drosophila species and clades. We wish to understand better the developmental time course of antennal development, segmentation, and specialization of various species with known courtship, aversive, and food specialization behaviors as well as the genetic underpinnings associated with these anatomical changes and provide novel genes of interest for understanding changes in the anatomy and physiology of Drosophila speciation.
The genetic network which underlies the changes in behavior also plays a role in the development and specification of olfactory sensory neuron (ORN) receptor expression, ORN number, size, and shape. Previous work on both forelimb and antennae development has shown the necessity of fruitless expression as well as additional factors like Dachshund, Distal-less, and Notch signaling pathways in the regulation of ORN position, antennal development, and segmentation [3,4,5]. Following upon these works current work in the lab aims to expand upon the knowledge of these systems by looking at the association of antennal development across various Drosophila species and clades. We wish to understand better the developmental time course of antennal development, segmentation, and specialization of various species with known courtship, aversive, and food specialization behaviors as well as the genetic underpinnings associated with these anatomical changes and provide novel genes of interest for understanding changes in the anatomy and physiology of Drosophila speciation.
[1] Zhao, et al. 2020. Sciences Advances. Chromatin-based reprogramming of a courtship regulator by concurrent pheromone perception and hormone signaling.
[2] Sethi, et al. 2019. Current Biology. Social Context Enhances Hormonal Modulation of Pheromone Detection in Drosophila.
[3] Li, et al. 2016. PLoS Genetics Social Context Enhances Hormonal Modulation of Pheromone Detection in Drosophila.
[4] Pan, et al. 2017. Science Reports. Patterns of transcriptional parallelism and variation in the developing olfactory system of Drosophila species.
[5] Ruiz-Losada, et al. 2018. Jour Dev Bio. Specification and Patterning of Drosophila Appendages
Seth O’Connor
I work on two projects in the Jones Lab. The first is aimed at understanding how genes evolve in Drosophila genomes. Every species has a subset of genes that exist only in that species and a broader set that exist only in closely related species (i.e., taxonomically restricted genes). My work is aimed at understanding how these genes evolve, how they are transcriptionally regulated, and how they function.
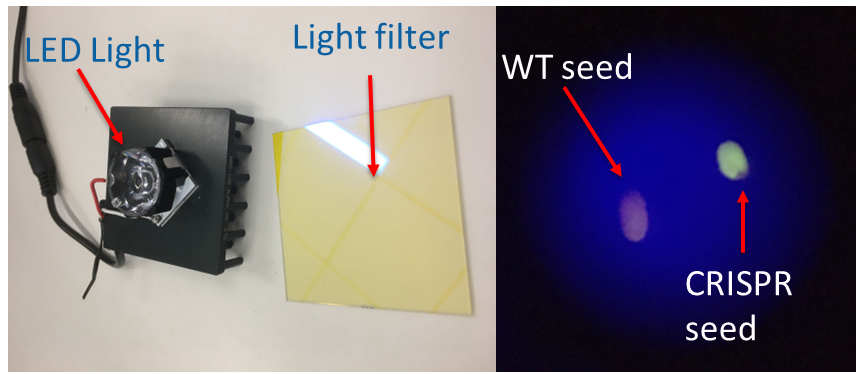 The second project I work on is aimed at developing an undergraduate-level, high-impact research project. This project utilizes the CRISPR system in Arabidopsis thaliana to perform cis-editing near high-functioning genes involved in senescence. Instead of knocking out genes with t-DNA insertions or overexpressing them with OE-vectors, we target and delete important regulatory sites outside of the gene of interest that regulate the activity of the gene of interest. By this method, we can generate non-transgenic plants with more fine-tuned alterations in gene activity. To make this project accessible at smaller primarily undergraduate colleges with fewer resources, a homemade fluorescence detection system for detecting our CRISPR vectors (with a YFP marker) was made for under $150 (see image). I am currently implementing this system here at UNC-CH as well as with collaborators at UNC-Pembroke.
The second project I work on is aimed at developing an undergraduate-level, high-impact research project. This project utilizes the CRISPR system in Arabidopsis thaliana to perform cis-editing near high-functioning genes involved in senescence. Instead of knocking out genes with t-DNA insertions or overexpressing them with OE-vectors, we target and delete important regulatory sites outside of the gene of interest that regulate the activity of the gene of interest. By this method, we can generate non-transgenic plants with more fine-tuned alterations in gene activity. To make this project accessible at smaller primarily undergraduate colleges with fewer resources, a homemade fluorescence detection system for detecting our CRISPR vectors (with a YFP marker) was made for under $150 (see image). I am currently implementing this system here at UNC-CH as well as with collaborators at UNC-Pembroke.
Wenbin (Bean) Zhou
Diego Urquia
My interests lie in using molecular tools to answer questions about the ecology, demography, and evolution of wildlife and plant populations, especially in the Galapagos Islands. I have experience in population genetics and bioinformatics, as well as in stable isotope analysis. At the Jones lab, I’m currently collaborating on the “Pastos Project” that is looking for the ‘key genes’ that lets different species of grass adapt to contrasting kinds of habitats, from the cold highlands of the Andes to the dry-hot Galapagos coasts. I’m also working on the “VFT Project”, where we use genomic tools for its application in the conservation of the endangered, Carolinas-endemic Venus Flytrap, the famous carnivore plant.
